Validation of an ELISA for quantitative determination of Mouse SARS-COV-2 IgG antibodies using clinical samples as a tool for vaccine development.
Authors: Krisha Jain, Atul Gadhave, Anitha Florance, Dr. Kalpesh Jain, Dharmendra Singh
Abstract:
Since the spread of SARS-CoV-2 in 2019 and the ensuing pandemic that led to a global halt, the race to develop an effective vaccine has been urgent and expedited. SARS-CoV-2 uses the receptor-binding domain (RBD) of its structural spike protein to engage with the receptor angiotensin-converting enzyme 2 (ACE2) on host cells, facilitating entry into the host. Thus, spike RBD has been an ideal candidate for vaccine development. To assess the efficacy of vaccines and to study disease mechanisms that help evaluate medical countermeasures, the use of animal models, especially mouse, has been the common practise for many decades. We have developed and validated a quantitative spike RBD antibody ELISA kit for mouse serum and plasma samples, useful for evaluating specific antibodies. The intention of this ELISA is to support vaccine and therapy development in pre-clinical stages. The assay employs an indirect sandwich technique. Antibodies present in the sample are bound by the SARS-COV-2 Spike-His Protein coated onto the plate microwells. Addition of a detection antibody forms an immune complex. Colour develops on addition of TMB substrate which is proportional to the amount of IgG antibodies present against Spike RBD protein. Commercially available mouse serum was spiked with SARS-COV-2 spike protein to evaluate the efficiency of the developed assay. The results showed high specificity for spike RBD protein and sensitivity was observed to be 12ng/ml. Recovery rates were between the accepted 80-120%. Inter and Intra assay precision and accuracy tests were performed with low, medium and high standards, that were determined to be <15>
Introduction:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive-sense single-stranded RNA virus that causes coronavirus infection (COVID-19), the respiratory illness responsible for the COVID-19 pandemic.[1][2]. Coronavirus has four proteins that build up its primary structure – the envelope protein, the spike protein, the nucleocapsid protein and the membrane protein.
Among all structural proteins of SARS-CoV-2, spike protein is the main antigenic component that is responsible for inducing host immune responses, neutralizing antibodies and/or protective immunity against virus infection [3]. The glycosylated spike (S) proteins covering the surface of SARS-COV-2 bind to receptor angiotensin-converting enzyme 2 (ACE-2) of the host cell and mediate the entry of the virus into the host. This binding of S protein to ACE-2 subsequently promotes entry into the host cell through TM protease serine 2 (TMPRSS2) and lysosomal proteases.
This important role of Spike protein in the causation of infection indicates that it is potential target for vaccine development, antibody-blocking therapy and small molecule inhibitors [4]. The role of spike protein (S protein) in receptor binding and membrane fusion also indicates that vaccines based on the spike protein could induce antibodies to block virus binding and fusion or neutralize virus infection [3].
Linlin Bao et. al. has confirmed the pathogenicity of SARS-CoV-2 in hACE2 mice. This mouse model of SARS-CoV-2 infection has been valuable for evaluating antiviral therapeutic agents and vaccines, as well as understanding the pathogenesis of COVID-19 [7].
J. Yang’s study of recombinant spike RBD protein as a protective immune response in mice, rabbits, and non-human primates results highlighted the importance of the spike RBD in SARS-CoV-2 vaccine design and provided a vaccine candidate to counteract COVID-19 [6]. Furthermore, a study on characterization of the receptor-binding domain for development of RBD vaccines where SARS-CoV RBD induced antibodies cross neutralised SARS-CoV-2 pseudo-virus infections, also indicated the relevance of the spike RBD.
Bringing of vaccines against the viral pathogen SARS-CoV-2 to the market for people around the world is essential, but rigorous studies are also required to determine the safety and efficacy of candidate vaccines. Assessing the potentiality of a candidate vaccine helps to evaluate the appropriate dosing regimen, to test its effect in generating neutralizing activity against SARS-COV-2 and to determine the immune pathways involved in the generation of the immune response, to provide the framework for the design of an effective vaccine. [6] So far, there are over 146 vaccine candidates registered with the WHO in various clinical trial phases, with several more in the pipeline.
ELISA based antibody tests for estimation of antibodies typically offer an easy, reliable method of detection. ELISA tests are easy to use and standardized, offering high levels of reproducibility and sensitivity. Mouse models can provide key insights into the pathogenic mechanisms of coronavirus disease and can serve as high-throughput preclinical evaluation platforms to identify highly performing antiviral agents and vaccines. These determinations have implications in vaccine development as well. As of today, there are only a small number of Mouse Anti-SARS-CoV-2 IgG ELISA Kits commercially available for determination of antibodies in mouse models. We believe that this assay showed usefulness in the determination of specific antibodies, imperative in the development of vaccines.
Objective:
In this study, we have developed a sensitive, specific enzyme linked-immunosorbent assay (ELISA) for measurement of IgG antibodies against mouse SARS-COV-2 spike RBD protein with the intention of aiding vaccine development and research.
Materials and Methods:
Materials Used
A DNA sequence encoding the SARS-CoV-2 (2019-nCoV) Spike Protein (RBD) (YP_009724390.1) (Arg319-Phe541) was expressed with a polyhistidine tag at the C-terminus. This protein was coated on CoStarTM high binding microtitre plates (Corning Cat. No. 9018), using in-house coating and blocking procedures to prepare the plates with proprietary solutions. The SARS-CoV-2 (2019-nCoV) Spike RBD antibody, a rabbit polyclonal antibody, was commercially sourced and used as a calibrator in the assay. The secondary detection antibody – Rabbit Anti-Mouse IgG(H+L) conjugated to HRP and TMB substrate was sourced from an in-house vendor. Wash buffer, sample diluent and other coating solutions were all prepared in-house.
Method
A step by step optimization and validation protocol was followed for the development of this assay. SARS-CoV-2 Spike RBD recombinant protein was tagged with -His, and then coated overnight onto microwells using proprietary coating and blocking solutions. A rabbit polyclonal SARS-CoV-2 Spike RBD was used as the standard / calibrator. The secondary antibody (Anti-Rabbit IgG) was conjugated to HRP to complete the sandwich format. For determining optimized concentrations of the coating antigen, and the detection antibody, various concentrations for each were tested until an optimal differentiated signal was obtained. To determine antibody titers, the assay was optimized using checkerboard titration experiments.
We followed an in-house development and optimization protocol for this assay. First, the samples and standards were pipetted into the pre-coated microwells. Antibodies to SARS-CoV-2 present in both the standards and samples bound to the protein antigen. After an incubation, the wells are washed and followed by addition of HRP-conjugated Detection IgG Antibody into each well and incubated to form a complex. After washing microwells in order to remove any non-specific binding, the substrate solution (TMB) is added to microwells and color develops proportionally to the amount of Anti-Mouse Anti-SARS-CoV-2 (2019-nCoV) in the sample. Color development is then stopped by addition of stop solution (made in house). Absorbance was measured at 450 nm using the Tecan Safire 2 reader.
Each assay step was optimized for optimal noise-to-signal ratio, working range, minimal % CV and relative error.
Since no commercial kit was available for result comparison, collaborative work was done with National Centre For Cell Science. SARS-CoV-2 Spike RBD antibody (Sino Biologicals Inc., USA (Cat No. 40592-MM57) was commercially sourced and provided to their laboratory. The antibody was serially diluted using sample diluent provided with the assay as per indicated concentration (90, 180, 360 and 720 ng/ml) and spiked at 1:1000 with commercially available mouse serum (Apollo Biosciences, Bengaluru, India). This spiked serum was used to run the assay for checking matrix interference and spike/recovery studies in duplicates. Mouse sera was measured with two replicates and two runs. Samples were measured using one lot of reagents. The recovery obtained was equated to our kit with comparable results.
Results:
Coefficient of Determination and Sensitivity
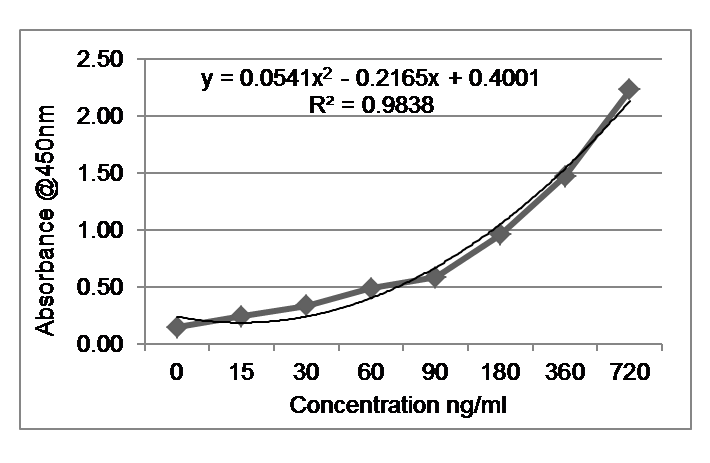
The assay was designed for 0 – 720 ng/ml as calibrator range.
Mean Absorbance of each set of duplicates of Samples and Standards were calculated. To determine the unknown Mouse Anti-SARS-CoV-2 Spike RBD IgG concentration, the average value of each standard on the Y-axis and the corresponding concentration of the standards on the X-axis was plotted. A polynomial regression (2nd order) curve-fit was drawn and unknowns were interpolated. The ELISA was optimized until recovery percentage of the unknown sample concentrations was between 90 – 110%.
The co-efficient of determination (R2) was >0.98. The sensitivity of this ELISA is 12 ng/ml.
Analytical Specificity
Mutations in the SARS-CoV-2 genome have been identified as the virus has spread, but no serologically unique strains have been described relative to the originally isolated virus (this research is limited at present). The antibodies purchased commercially for the development of this ELISA are specific to spike RBD of the SARS-CoV-2 virus, with thorough specificity testing performed on the manufacturers end.
Precision
Precision is defined as the percent coefficient of variation (% CV) i.e. the average standard deviation divided by the average mean of all samples and multiplied by 100. Assay precision was determined by both intra (n=5 assays) and inter assay (n=5 assays) reproducibility on two pools. While actual precision may vary from laboratory to laboratory and technician to technician, it is recommended that all operators achieve precision below these design goals before reporting results.
Recovery
All data met our acceptance criteria for % CV and 95% (CI) Confidence Intervals for % CV.
Recoveries for serum was observed between 90 – 110%.
Discussion:
Typically, mouse models are the most commonly used animal model during vaccine developments. A number of transgenic and knock-in mouse models, as well as viral vector-mediated hACE2 delivery systems, have been developed to understand disease pathogenesis for evaluation of vaccines and antivirals. Small animal models also allow us to better characterize the immune response of vaccine combinations or immunization regimens.
With the increased infectiousness of the new variants – most importantly the Delta variant of the virus, the use of a permissive small animal model facilitates broad applications and thorough research. Hence, an accurate, specific technique for detecting SARS-CoV-2 Spike RBD protein in samples where mouse models are used for vaccine evaluation is necessary. ELISA is a reliable, sensitive and easy-to-use technology which can be used and also there is a need for the development of kit as there very few commercially kits available in the market to understand the vaccine-induced protective immunity. Therefore, the development of this ELISA holds an important place in the research and development space with mouse models. The use of a commercially available ELISA for the quantification of these antibodies in mouse sera ensures a pre-validated, accurate protocol with standardized, accurate results.
Conclusion:
The GENLISA™ SARS-CoV-2 Spike RBD Antibody Quantitative ELISA (Mouse) developed and validated as above gives accurate results. The method used above is fast, reproducible and reliable, and results from our data shows that the use of monoclonal antibodies specific for SARS-COV-2 Spike RBD resulted in a highly specific and sensitive ELISA test with sensitivity of 12ng/ml. The data above also shows that the kit can detect the antibody titres post vaccination in mouse, which is imperative in dosage studies as well. Thus, we can reasonably conclude that the Mouse SARS-CoV-2 Spike RBD antibodies ELISA discussed here is a valuable addition to the testing repertoire of vaccine developers.
References:
-
“CoV2020”. GISAID EpifluDB. Archived from the original on 12 January 2020. Retrieved 12 January 2020.^
-
Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Wanbo Tai, Lei He, Xiujuan Zhang, Jing Pu, Denis Voronin, Shibo Jiang, Yusen Zhou & Lanying Du,Cellular & Molecular Immunology volume 17, pages613–620 (2020)
-
Yang, J., Wang, W., Chen, Z. et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 586, 572–577 (2020). https://doi.org/10.1038/s41586-020-2599-8
-
Bao, L., Deng, W., Huang, B. et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583, 830–833 (2020). https://doi.org/10.1038/s41586-020-2312-y
-
Pickering S, Betancor G, Galão RP, Merrick B, Signell AW, Wilson HD, et al. (2020) Comparative assessment of multiple COVID-19 serological technologies supports continued evaluation of point-of-care lateral flow assays in hospital and community healthcare settings. PLoS Pathog 16(9): e1008817. https://doi.org/10.1371/journal.ppat.1008817
-
Yin-Feng Kang, Cong Sun, Zhen Zhuang, Run-Yu Yuan, Qingbing Zheng, Jiang-Ping Li, Ping-Ping Zhou, Xin-Chun Chen, Zhe Liu, Xiao Zhang, Xiao-Hui Yu, Xiang-Wei Kong, Qian-Ying Zhu, Qian Zhong, Miao Xu, Nan-Shan Zhong, Yi-Xin Zeng, Guo-Kai Feng, Changwen Ke, Jin-Cun Zhao, and Mu-Sheng Zeng
ACS Nano 2021 15 (2), 2738-2752, DOI: 10.1021/acsnano.0c08379
-
Kang, Y. F., Sun, C., Zhuang, Z., Yuan, R. Y., Zheng, Q., Li, J. P., Zhou, P. P., Chen, X. C., Liu, Z., Zhang, X., Yu, X. H., Kong, X. W., Zhu, Q. Y., Zhong, Q., Xu, M., Zhong, N. S., Zeng, Y. X., Feng, G. K., Ke, C., . . . Zeng, M. S. (2021). Rapid Development of SARS-CoV-2 Spike Protein Receptor-Binding Domain Self-Assembled Nanoparticle Vaccine Candidates. ACS Nano, 15(2), 2738–2752. https://doi.org/10.1021/acsnano.0c08379
-
Huo, J., Le Bas, A., Ruza, R.R. et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat Struct Mol Biol 27, 846–854 (2020). https://doi.org/10.1038/s41594-020-0469-6



