Introduction: The J2 anti-dsRNA IgG2a monoclonal antibody (Schonborn et al. 1991) has become the gold standard in dsRNA detection. It was used initially for the study of plant viruses, but since the seminal paper of Weber et al. in 2006, where J2 was used to show that all the positive strand RNA viruses tested produced copious amounts of dsRNA in infected cells, this antibody has been used extensively in a wide range of systems, as documented in over 200 scientific publications. J2 can be used to detect dsRNA intermediates of viruses as diverse as Hepatitis C virus, Dengue virus, rhinovirus, Chikungunya virus, Rabies virus, Polio virus, Classic swine fever virus, Brome mosaic virus and many more in cultured cells and also in fixed paraffin-embedded histological samples.
J2 has been used to elucidate how anti-viral responses are initiated, what counter-strategies viruses have adopted to avoid them, and to explore the viral life cylce by enabling ultrastructiural localisation studies of viral nucleic acid replication sites (Welsch et al., 2009 & Knoops et al., 2011). J2 has also been recommended as a diagnostic tool to detect whether an unkown pathogen is bacterial or viral in nature (Richardson et al., 2010). Recently J2 has also been used to monitor the removal of dsRNA from in vitro synthethised mRNA preparations that may have potential use in gene therapy (Kariko et al., 2011). J2 has been used successfully in various immunocapture methods, such as ELISA.
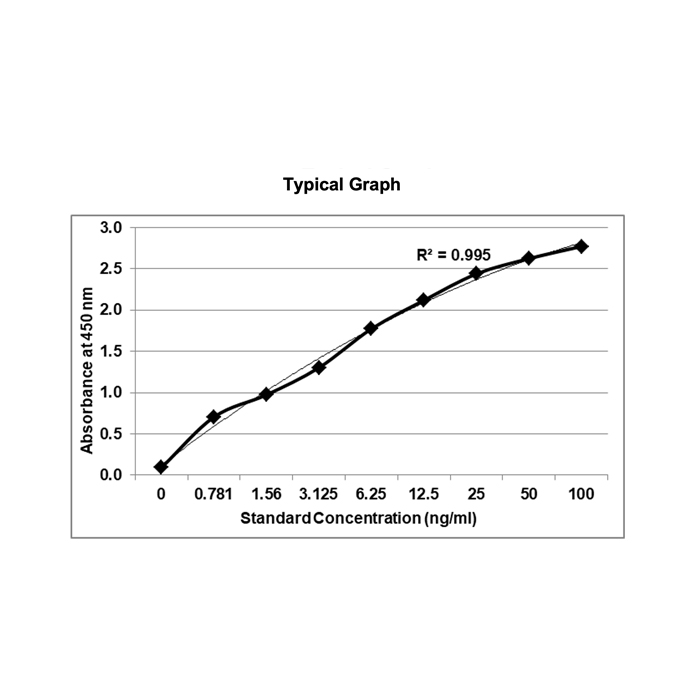
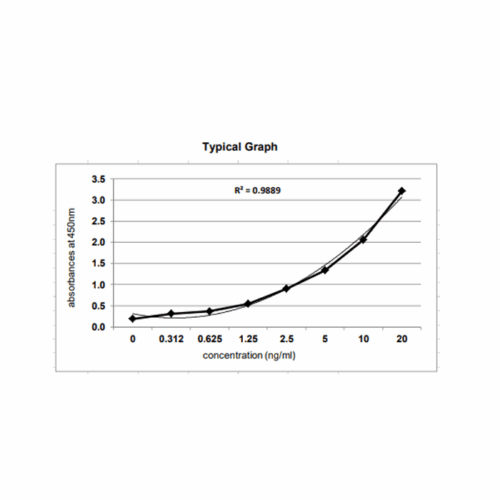
Assay Principle: The KRIBIOLISA Double-Stranded RNA (dsRNA) ELISA employs the quantitative sandwich enzyme immunoassay technique. It is based on the use of two double-stranded RNA (dsRNA)-specific monoclonal antibodies which allows sensitive and selective detection of dsRNA molecules (& gt;=40 bp), independent of their nucleotide composition and sequence. Antibodies to dsRNA (J2) are pre-coated onto microwells. Samples and standards are pipetted into microwells and are bound by the capture antibody. Then, a HRP (horseradish peroxidase) conjugated Anti-dsRNA (K1) is pipetted and incubated. After washing microwells in order to remove any non-specific binding, the ready to use substrate solution (TMB) is added to microwells and color develops proportionally to the amount of dsRNA in the sample. Color development is then stopped by addition of stop solution. Absorbance is measured at 450 nm.
![]() If you have published a paper by using any of our ELISA since 01/01/2023, kindly fill out the “Krishgen Publication Reward Application Form” with complete information and send it by at email: kbiinfo@krishgen.com, with the subject “Krishgen Publication Reward”. We will get back to you with the Amazon / Krishgen Credit Reward after we confirm it ASAP!
If you have published a paper by using any of our ELISA since 01/01/2023, kindly fill out the “Krishgen Publication Reward Application Form” with complete information and send it by at email: kbiinfo@krishgen.com, with the subject “Krishgen Publication Reward”. We will get back to you with the Amazon / Krishgen Credit Reward after we confirm it ASAP!